When experiments on peas were repeated using other traits in other plants, it was found that sometimes the F1 had a phenotype that did not resemble either of the two parents and was in between the two. The inheritance of flower colour in the dog flower (snapdragon or Antirrhinum sp.) is a good example to understand incomplete dominance. In a cross between true-breeding red-flowered (RR) and true-breeding white-flowered plants (rr), the F1 (Rr) was pink (Figure 5.6). When the F1 was self-pollinated the F2 resulted in the following ratio 1 (RR) Red: 2 (Rr) Pink: 1 (rr) White. Here the genotype ratios were exactly as we would expect in any mendelian monohybrid cross, but the phenotype ratios had changed from the 3:1 dominant : recessive ratio. What happened was that R was not completely dominant over r and this made it possible to distinguish Rr as pink from RR (red) and rr (white) .
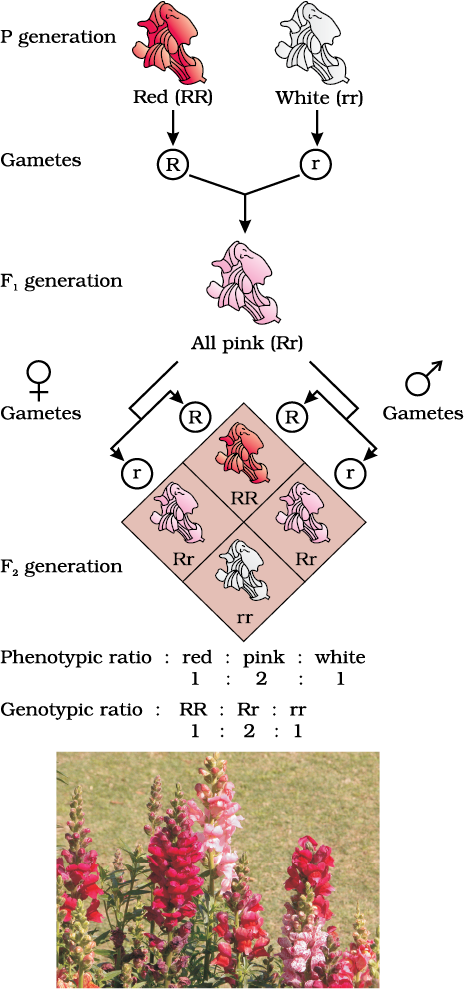
Figure 5.6 Results of monohybrid cross in the plant Snapdragon, where one allele is incompletely dominant over the other allele
Explanation of the concept of dominance: What exactly is dominance? Why are some alleles dominant and some recessive? To tackle these questions, we must understand what a gene does. Every gene, as you know by now, contains the information to express a particular trait. In a diploid organism, there are two copies of each gene, i.e., as a pair of alleles. Now, these two alleles need not always be identical, as in a heterozygote. One of them may be different due to some changes that it has undergone (about which you will read further on, and in the next chapter) which modifies the information that particular allele contains.
Let’s take an example of a gene that contains the information for producing an enzyme. Now there are two copies of this gene, the two allelic forms. Let us assume (as is more common) that the normal allele produces the normal enzyme that is needed for the transformation of a substrate S. Theoretically, the modified allele could be responsible for production of –
(i) the normal/less efficient enzyme, or
(ii) a non-functional enzyme, or
(iii) no enzyme at all
In the first case, the modified allele is equivalent to the unmodified allele, i.e., it will produce the same phenotype/trait, i.e., result in the transformation of substrate S. Such equivalent allele pairs are very common. But, if the allele produces a non-functional enzyme or no enzyme, the phenotype may be effected. The phenotype/trait will only be dependent on the functioning of the unmodified allele. The unmodified (functioning) allele, which represents the original phenotype is the dominant allele and the modified allele is generally the recessive allele. Hence, in the example above the recessive trait is seen due to non-functional enzyme or because no enzyme is produced.