You know that plasmids and bacteriophages have the ability to replicate within bacterial cells independent of the control of chromosomal DNA. Bacteriophages because of their high number per cell, have very high copy numbers of their genome within the bacterial cells. Some plasmids may have only one or two copies per cell whereas others may have 15-100 copies per cell. Their numbers can go even higher. If we are able to link an alien piece of DNA with bacteriophage or plasmid DNA, we can multiply its numbers equal to the copy number of the plasmid or bacteriophage. Vectors used at present, are engineered in such a way that they help easy linking of foreign DNA and selection of recombinants from non-recombinants.
The following are the features that are required to facilitate cloning into a vector.
(i) Origin of replication (ori) : This is a sequence from where replication starts and any piece of DNA when linked to this sequence can be made to replicate within the host cells. This sequence is also responsible for controlling the copy number of the linked DNA. So, if one wants to recover many copies of the target DNA it should be cloned in a vector whose origin support high copy number.
(ii) Selectable marker : In addition to ‘ori’, the vector requires a selectable marker, which helps in identifying and eliminating non-transformants and selectively permitting the growth of the transformants. Transformation is a procedure through which a piece of DNA is introduced in a host bacterium (you will study the process in subsequent section). Normally, the genes encoding resistance to antibiotics such as ampicillin, chloramphenicol, tetracycline or kanamycin, etc., are considered useful selectable markers for E. coli. The normal E. coli cells do not carry resistance against any of these antibiotics.
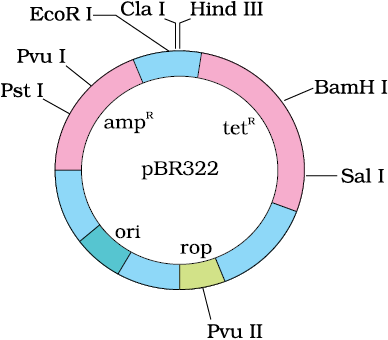
Figure 11.4 E. coli cloning vector pBR322 showing restriction sites
(iii) Cloning sites: In order to link the alien DNA, the vector needs to have very few, preferably single, recognition sites for the commonly used restriction enzymes. Presence of more than one recognition sites within the vector will generate several fragments, which will complicate the gene cloning (Figure 11.4). The ligation of alien DNA is carried out at a restriction site present in one of the two antibiotic resistance genes. For example, you can ligate a foreign DNA at the BamH I site of tetracycline resistance gene in the vector pBR322. The recombinant plasmids will lose tetracycline resistance due to insertion of foreign DNA but can still be selected out from non-recombinant ones by plating the transformants on tetracycline containing medium. The transformants growing on ampicillin containing medium are then transferred on a medium containing tetracycline. The recombinants will grow in ampicillin containing medium but not on that containing tetracycline. But, non- recombinants will grow on the medium containing both the antibiotics. In this case, one antibiotic resistance gene helps in selecting the transformants, whereas the other antibiotic resistance gene gets ‘inactivated due to insertion’ of alien DNA, and helps in selection of recombinants.
Selection of recombinants due to inactivation of antibiotics is a cumbersome procedure because it requires simultaneous plating on two plates having different antibiotics. Therefore, alternative selectable markers have been developed which differentiate recombinants from non-recombinants on the basis of their ability to produce colour in the presence of a chromogenic substrate. In this, a recombinant DNA is inserted within the coding sequence of an enzyme, β-galactosidase. This results into inactivation of the gene for synthesis of this enzyme, which is referred to as insertional inactivation. The presence of a chromogenic substrate gives blue coloured colonies if the plasmid in the bacteria does not have an insert. Presence of insert results into insertional inactivation of the β-galactosidase gene and the colonies do not produce any colour, these are identified as recombinant colonies.
(iv) Vectors for cloning genes in plants and animals : You may be surprised to know that we have learnt the lesson of transferring genes into plants and animals from bacteria and viruses which have known this for ages – how to deliver genes to transform eukaryotic cells and force them to do what the bacteria or viruses want. For example, Agrobacterium tumifaciens, a pathogen of several dicot plants is able to deliver a piece of DNA known as ‘T-DNA’ to transform normal plant cells into a tumor and direct these tumor cells to produce the chemicals required by the pathogen. Similarly, retroviruses in animals have the ability to transform normal cells into cancerous cells. A better understanding of the art of delivering genes by pathogens in their eukaryotic hosts has generated knowledge to transform these tools of pathogens into useful vectors for delivering genes of interest to humans. The tumor inducing (Ti) plasmid of Agrobacterium tumifaciens has now been modified into a cloning vector which is no more pathogenic to the plants but is still able to use the mechanisms to deliver genes of our interest into a variety of plants. Similarly, retroviruses have also been disarmed and are now used to deliver desirable genes into animal cells. So, once a gene or a DNA fragment has been ligated into a suitable vector it is transferred into a bacterial, plant or animal host (where it multiplies).